Research
Research
In our work we use and develop methods of statistical and mathematical physics to attack important biological questions. We are interested in applications but also devote a significant part of our work to advancing the fundamental theoretical basis. Our group has a remarkable record of collaborative work, both, with theorists and experimentalist.
Biofilm as a model of a multicellular organism
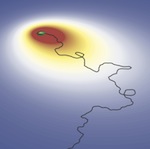
Biofilms are dense multicellular communities of bacteria that form on surfaces. Biofilms are widespread in nature and implicated in many acute medical and industrial problems. We now turned to the early stages of biofilm development and want to understand how biofilms form. For that we consider the self-assembly of N.gonorrhoeae micro-colonies. These bacteria cause one of the most common sexually transmitted diseases, gonorrhoea, where the micro-colonies represent the infectious unit of the disease. In a series of work we demonstrated that cells use long retractile filaments called pili to attach and move on surfaces, attach to other cells and self-assemble into spherical colonies containing thousands of cells. Here we applied a variety of statistical physics approaches from modeling to the analytical framework of kinetic and hydrodynamic descriptions. Recently we were able to create an in silico model of the bacterialcolony with explicit pili dynamics and interaction forces that recapitulates our experimental observations on various space and time scales. We could show that the pili forces were responsible for the emergence of heterogeneous cell motility within large micro-colonies of essentially identical cells. The presented figure illustrates the cross-section of the micro-colony where the mutant cells (red) that are not able to retract pili get segregated to the outside of the wild-type cells (yellow). This work is a collaboration with a group of Nicolas Biais.
From chromosomal loops to Brownian bridges, ASEP and single file diffusion
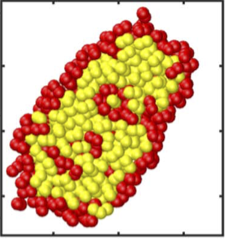
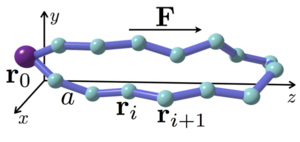
Recently we investigated the problem of chromosome alignment for recombination during meiosis. This distinct biological problem appeared to be linked to a gold-mine of statistical physics problems. We first could reduce the original problem to finding the conformation of pinned polymer loops under constant external force (see figure). This in turn can be formulated in the language of random walks and Brownian bridges. Surprisingly the found analytical solution relied on the Fermi-Dirac statistics. To study the relaxation dynamics of such polymer loops we used the mapping to the particle system of asymmetric exclusion process (ASEP) which in turn can be solved by using the Bethe ansatz methodology. Further analogy can be drawn to the single-file diffusion in the finite domain with reflecting boundaries. This work is in progress and it shows that very seemingly distant approaches of statistical physics can be used to analytically attack problems motivated by concrete biological questions.
Phase transitions in cell-cytoplasm at energy depletion and desiccation
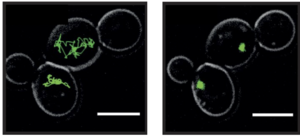
In a recent collaborative effort (with S. Alberti, MPI-CBG, and J. Guck, TU Dresden) we have discovered that single-cell organisms such as yeast or amoeba can survive energy depletion by transforming their cytoplasm from a fluid-like state to a solid-like state. We used several techniques such as passive microrheology, AFM measurements, and real-time deformability cytometry to demonstrate that cells transition to a state with pronounced elastic properties. Figure shows the trajectories of tracer particles in budding yeast cells in two different states: high exploration of cytoplasm in the fluid-like state (left) and almost no motion in the solid-like state (right). It was shown that the major cause of this transition is the drop in intracellular pH which leads to a widespread self-assembly of cytosolic proteins.
We have planned an extensive program to further develop this line of research. Our collaboration extended by group of T. Kurzchalia, MPI-CBG, will focus on the related transition of the cells and organisms (C.elegans worm) during desiccation. It was previously shown in plant seeds that the upregulated production of special sugars (sucrose in plants and trehalose in C.elegance) leads to the glass transition in seeds at the removal of water. What is the physical mechanism of sugars conservative role remains poorly understood and we would like to contribute to answering this challenging question.
Transcription activation in early developing embryo
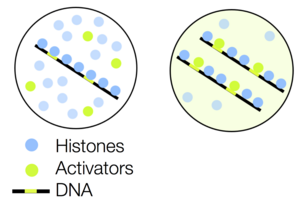
In plants and animals, at the very first early stages of embryo development the genome is transcriptionally inactive. Only after several cell division cycles genome gets activated and takes over the control over the development. Together with our experimental collaborators (group of N. Vastenhouw, MPI-CBG Dresden) we could show that the silencing of the transcription is achieved via histone proteins competing with transcription activation factors. The chromatin bound to histones and forming nucleosomes is inaccessible for transcription factors. As DNA keeps replicating, the histones get depleted and eventually loose the competition, allowing for the onset of transcription. We now exploit the onset of genome activation to investigate the effect of transcriptional activity on the chromatin structure.
Lévy walks & Anomalous Diffusion
Random walk models are excellent analytical tools for addressing a number of important questions of statistical physics: various transport processes including normal and anomalous diffusion, aging, memory effects, finite or random velocity of random walkers, and transport in active media. One of the most advanced models is the Lévy walk model, which we have recently reviewed (with S. Denisov). We continue to explore this model by extending its theory to higher dimensions.
Chemotaxis
Chemotaxis is the mechanism allowing cells to react on the concentration of chemicals such as nutrients or signaling molecules. Cells or bacteria are able to find food, each other or escape dangerous conditions by adjusting their behavior in accordance with environmental conditions. We study how individual cells use their motility patterns to localize the source of attractant in the most efficient way. We also look at how many cells can agglomerate by producing signaling molecules that attract their neighbors (in collaboration with S. Denisov, C. Beta, and H.Stark).