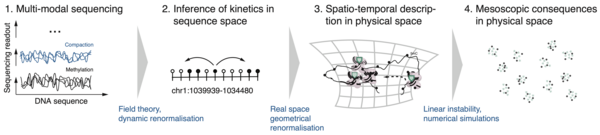
Biological systems continuously use energy to build complex structures in space and time. Examples are the complex interactions between molecules among each other and with the DNA or the self-organisation of cells into complex organs. Understanding the rules governing such systems is pivotal for developing therapeutic strategies for diseases related to the breaking of these rules. Together with experimental collaborators researchers in our group apply methods from theoretical physics, data science and artificial intelligence to understand the working principles underlying biological systems.